Plotting atomic partial charges from Critic2 output-files¶
Reading the output files¶
To plot the partial charges from the Critic2 output the function read_critic2_stdout from the io sub-package can be used to parse the elements and populations from the output file:
[1]:
from aim2dat.io import read_critic2_stdout
Personally, I like to define a dictionary which contains the data:
[2]:
partial_data = {}
partial_data["ZIF_8_Cl_struc"] = read_critic2_stdout("./files/pc_critic2/ZIF-8-Cl-struc-critic2.out")[
"partial_charges"
]
partial_data["ZIF_8_Cl_comp"] = read_critic2_stdout("./files/pc_critic2/ZIF-8-Cl-comp-critic2.out")[
"partial_charges"
]
partial_data["ZIF_8_Cl_imi"] = read_critic2_stdout("./files/pc_critic2/ZIF-8-Cl-imi-critic2.out")[
"partial_charges"
]
[3]:
partial_data["ZIF_8_Br_struc"] = read_critic2_stdout("./files/pc_critic2/ZIF-8-Br-struc-critic2.out")[
"partial_charges"
]
partial_data["ZIF_8_Br_comp"] = read_critic2_stdout("./files/pc_critic2/ZIF-8-Br-comp-critic2.out")[
"partial_charges"
]
partial_data["ZIF_8_Br_imi"] = read_critic2_stdout("./files/pc_critic2/ZIF-8-Br-imi-critic2.out")[
"partial_charges"
]
The output of the function is a list containing dictionaries of elements and their population
[4]:
partial_data["ZIF_8_Br_imi"]
[4]:
[{'element': 'C', 'population': 3.04080751},
{'element': 'N', 'population': 6.20870328},
{'element': 'C', 'population': 3.6766472},
{'element': 'C', 'population': 3.60243296},
{'element': 'N', 'population': 6.04921432},
{'element': 'Br', 'population': 7.02320405},
{'element': 'H', 'population': 0.938178293},
{'element': 'H', 'population': 0.919212183},
{'element': 'H', 'population': 0.541557784}]
Optional:
The elements of the example data contain different symmetries, and thus the calculated charge can be arranged in a favorable manner. Therefore, a dictonary of elements combined with a list of indicies can be defined:
elementscan be split in for example {“H1”:[6,7], “H2”:[8]}elementscan be combined {“conjugated_network”: [[0],[2,3],[6,7],[8]]}. The mean of each element will be calculated and summed
[5]:
index_struc = {
"X": [42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53],
"Zn": [0, 1, 2, 3, 4, 5],
"N": [
54,
55,
56,
57,
58,
59,
61,
62,
63,
66,
67,
68,
69,
70,
71,
72,
76,
77,
60,
64,
65,
73,
74,
75,
],
"conjugated_network": [
[6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17],
[
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33,
34,
35,
36,
37,
38,
39,
40,
41,
],
[
78,
79,
80,
81,
82,
83,
84,
85,
86,
87,
88,
89,
90,
91,
92,
93,
94,
95,
96,
97,
98,
99,
100,
101,
],
],
}
index_comp = {
"Zn": [0],
"X": [9, 25, 10, 27],
"N": [3, 6, 4, 5, 17, 18, 19, 26],
"conjugated_network": [
[1, 2, 13, 14, 22, 23, 24, 29],
[11, 20, 12, 21],
[7, 8, 15, 16, 28, 30, 31, 32],
[33, 34],
],
}
index_imi = {"conjugated_network": [[0], [2, 3], [6, 7], [8]], "N": [1, 4], "X": [5]}
Since the population depends on the number of valence electrons, we need to supply this information. Each element needs to be defined:
[6]:
valence_electrons = {"Zn": 12, "H": 1, "C": 4, "N": 5, "Br": 7, "Cl": 7}
Initialize the plot class¶
Now the PartialChargesPlot class in the plots sub-package is used to visualize the partial charges. Additional attributes can be set to show and store the plot:
[7]:
from aim2dat.plots.partial_charges import PartialChargesPlot
pc_plot = PartialChargesPlot()
pc_plot.store_path = "."
pc_plot.store_plot = False
pc_plot.show_plot = True
Importing data into class¶
The partial charge is imported with the import_partial_charges function. All data sets have its own data_label to distinguish them. Additionally, these parameters need to be set:
plot_label: groups certain datasets into on classx_label: distinguishes the datasets into different groups e.g. different functionalization or structurescustom_kind_dict: optional if not defined, the mean of each element will be calculated
[8]:
pc_plot.import_partial_charges(
data_label="ZIF_8_Cl_struc",
partial_charges=partial_data["ZIF_8_Cl_struc"],
valence_electrons=valence_electrons,
plot_label="Cl",
x_label="structure",
custom_kind_dict=index_struc,
)
pc_plot.import_partial_charges(
data_label="ZIF_8_Cl_comp",
partial_charges=partial_data["ZIF_8_Cl_comp"],
valence_electrons=valence_electrons,
plot_label="Cl",
x_label="complex",
custom_kind_dict=index_comp,
)
pc_plot.import_partial_charges(
data_label="ZIF_8_Cl_imi",
partial_charges=partial_data["ZIF_8_Cl_imi"],
valence_electrons=valence_electrons,
plot_label="Cl",
x_label="imidazole",
custom_kind_dict=index_imi,
)
[9]:
pc_plot.import_partial_charges(
data_label="ZIF_8_Br_struc",
partial_charges=partial_data["ZIF_8_Br_struc"],
valence_electrons=valence_electrons,
plot_label="Br",
x_label="structure",
custom_kind_dict=index_struc,
)
pc_plot.import_partial_charges(
data_label="ZIF_8_Br_comp",
partial_charges=partial_data["ZIF_8_Br_comp"],
valence_electrons=valence_electrons,
plot_label="Br",
x_label="complex",
custom_kind_dict=index_comp,
)
pc_plot.import_partial_charges(
data_label="ZIF_8_Br_imi",
partial_charges=partial_data["ZIF_8_Br_imi"],
valence_electrons=valence_electrons,
plot_label="Br",
x_label="imidazole",
custom_kind_dict=index_imi,
)
Plotting data¶
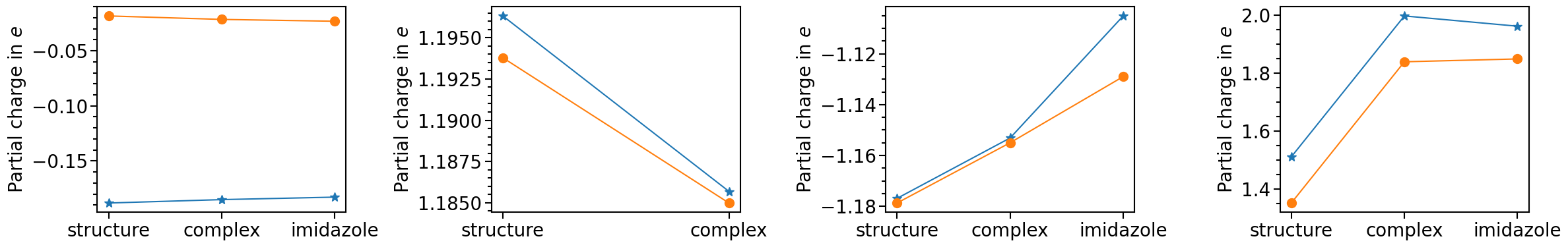
Data sets chosen by their data_label are plotted with the function plot.
[10]:
plot = pc_plot.plot(
data_labels=[
"ZIF_8_Cl_struc",
"ZIF_8_Cl_comp",
"ZIF_8_Cl_imi",
"ZIF_8_Br_struc",
"ZIF_8_Br_comp",
"ZIF_8_Br_imi",
]
)
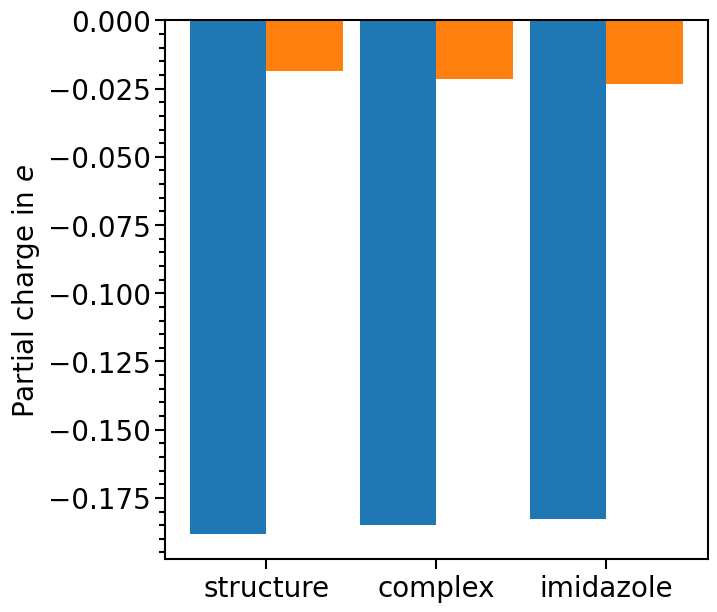
The plot typ can be set via the attribute pc_plot_type. The options are "scatter" or "bar":
[11]:
pc_plot.pc_plot_type = "bar"
plot = pc_plot.plot(
data_labels=[
"ZIF_8_Cl_struc",
"ZIF_8_Cl_comp",
"ZIF_8_Cl_imi",
"ZIF_8_Br_struc",
"ZIF_8_Br_comp",
"ZIF_8_Br_imi",
]
)
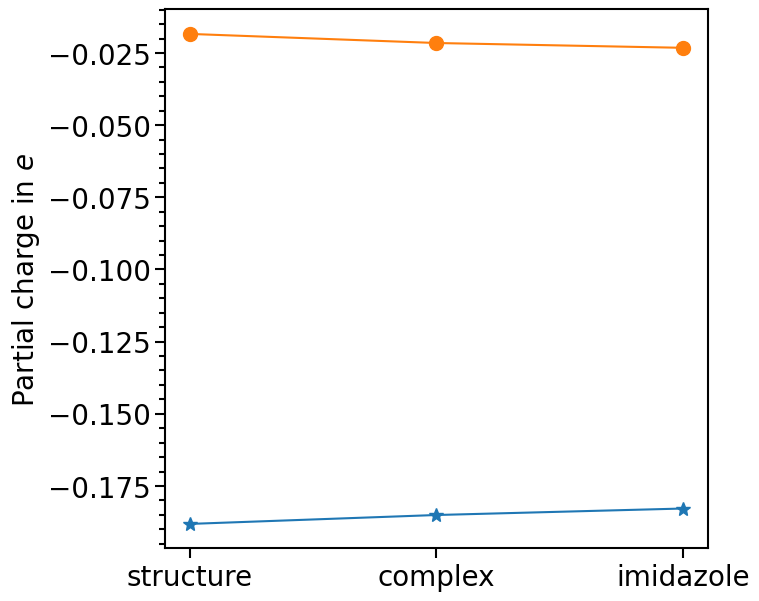
ratio: modifies the size of the figure in inchsubplot_tight_layout: adjusts thehspaceandwspaceautomaticallysubplot_ncolsandsubplot_nrows: shows multiple plots with the differentelements
[12]:
pc_plot.pc_plot_type = "scatter"
pc_plot.ratio = 24, 4
pc_plot.subplot_tight_layout = True
pc_plot.subplot_ncols, pc_plot.subplot_nrows = 4, 1
plot = pc_plot.plot(
data_labels=[
"ZIF_8_Cl_struc",
"ZIF_8_Cl_comp",
"ZIF_8_Cl_imi",
"ZIF_8_Br_struc",
"ZIF_8_Br_comp",
"ZIF_8_Br_imi",
]
)
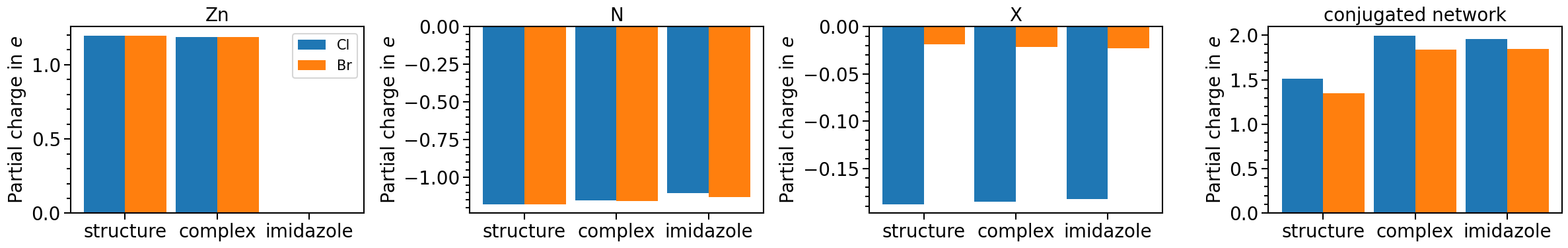
pc_plot_order: arranges theelementsin given order for subplotsshow_legend: adds a legend. Each subplot can be set individually but if(True, False)is set, first plot is set toTrue, rest is set toFalseplot_title: adds a title to each subplot
[13]:
pc_plot.pc_plot_type = "bar"
pc_plot.show_legend = (True, False)
pc_plot.ratio = 24, 4
pc_plot.subplot_tight_layout = True
pc_plot.subplot_ncols, pc_plot.subplot_nrows = 4, 1
pc_plot.pc_plot_order = ["Zn", "N", "X", "conjugated_network"]
plot = pc_plot.plot(
data_labels=[
"ZIF_8_Cl_struc",
"ZIF_8_Cl_comp",
"ZIF_8_Cl_imi",
"ZIF_8_Br_struc",
"ZIF_8_Br_comp",
"ZIF_8_Br_imi",
],
plot_title=["Zn", "N", "X", "conjugated network"],
)